Selectivity and Resolving Power of Hydrophobic Interaction Chromatography Targeting the Separation of Monoclonal Antibody Variants
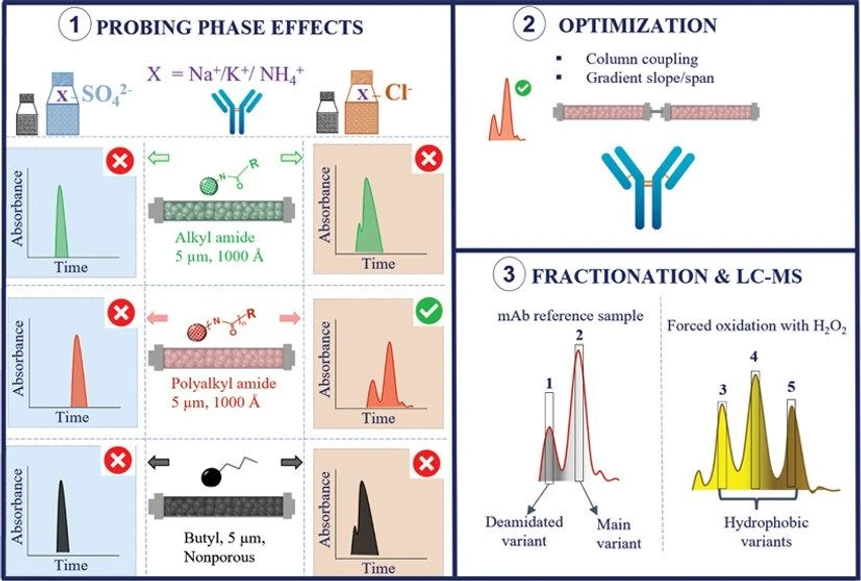
- Photo: Anal. Chem. 2024, 96, 3, 1121-1128: graphical abstract.
In the research article published in ACS Analytical Chemistry journal the international team of researchers from Vrije Universiteit Brussel, University of Salzburg, University of Natural Resources and Life Sciences Vienna, CAP Chromatography Consulting, California, and University of Salzburg presented a comprehensive investigation of the mechanistic understanding of retention and selectivity in hydrophobic interaction chromatography.
This study delves into the mechanisms behind retention and selectivity in hydrophobic interaction chromatography (HIC) for profiling molecular variants of trastuzumab. By evaluating three column chemistries—butyl, alkylamide, and multialkylamide—it highlights the impact of column hydrophobicity, surface area, and the role of chloride ions in enhancing chromatographic selectivity. The multialkylamide column demonstrated improved selectivity due to protein intercalation and steric effects. Furthermore, LC–MS/MS peptide mapping revealed that chloride in the mobile phase facilitated site-specific deamidation and oxidation, offering insights into structural changes impacting trastuzumab's retention behavior.
The original article
Selectivity and Resolving Power of Hydrophobic Interaction Chromatography Targeting the Separation of Monoclonal Antibody Variants
Raphael Ewonde Ewonde, Katharina Böttinger, Jelle De Vos, Nico Lingg, Alois Jungbauer, Christopher A. Pohl, Christian G. Huber, Gert Desmet, and Sebastiaan Eeltink
Analytical Chemistry 2024 96 (3), 1121-1128
DOI: 10.1021/acs.analchem.3c04011
licensed under CC-BY 4.0
Selected sections from the article follow.
Abstract
This study presents a comprehensive investigation of the mechanistic understanding of retention and selectivity in hydrophobic interaction chromatography. It provides valuable insights into crucial method-development parameters involved in achieving chromatographic resolution for profiling molecular variants of trastuzumab. Retention characteristics have been assessed for three column chemistries, i.e., butyl, alkylamide, and long-stranded multialkylamide ligands, while distinguishing column hydrophobicity and surface area. Salt type and specifically chloride ions proved to be the key driver for improving chromatographic selectivity, and this was attributed to the spatial distribution of ions at the protein surface, which is ion-specific. The effect was notably more pronounced on the multialkylamide column, as proteins intercalated between the multiamide polymer strands, enabling steric effects. Column coupling proved to be an effective approach for maximizing resolution between molecular variants present in the trastuzumab reference sample and trastuzumab variants induced by forced oxidation. Liquid chromatography–mass spectrometry (LC–MS)/MS peptide mapping experiments after fraction collection indicate that the presence of chloride in the mobile phase enables the selectivity of site-specific deamidation (N30) situated at the heavy chain. Moreover, site-specific oxidation of peptides (M₂₅₅, W₄₂₀, and M₄₃₁) was observed for peptides situated at the Fc region close to the CH2–CH3 interface, previously reported to activate unfolding of trastuzumab, increasing the accessible surface area and hence resulting in an increase in chromatographic retention.
Experimental Section
HIC Instrumentation and Conditions
HIC experiments were conducted using an Ultimate 3000 bioinert HPLC system (Thermo Fisher Scientific, Germering, Germany) composed of a low-pressure gradient pump, a well-plate autosampler enabling inline split-loop injections, a forced-air column oven, and a diode-array UV detector. Chromeleon software (version 7.2.10) was used for system control and data management. The injection volume was set to 5 μL. The temperature of the well-plate sampler was set at 6 °C and the column oven was maintained at 30 °C. A 2.5 μL UV flow cell was used at 210 and 280 nm with a data collection rate of 5 Hz and a response time of 1 s. The columns used in this study were 4.6 mm I.D. × 100 mm packed with 5 μm porous (1000 Å) silica particles functionalized with alkylamide chemistries (MabPac HIC-10 and MabPac HIC-20) or packed with 5 μm nonporous polymer particles functionalized with butyl chemistry (HIC-butyl) obtained from Thermo Fisher Scientific (Sunnyvale). HIC analyses were generally performed in gradient mode by applying inverse salt gradients starting at 2 M (NH4)2SO4 or 4 M NaCl in 50 mM phosphate buffer and decreasing the salt concentration linearly in time. All mobile phases were filtered through a 0.22 μm filter (Millipore, Darmstadt, Germany).
RP-HPLC-MS/MS Peptide Profiling
HPLC-MS/MS experiments were performed using an Ultimate 3000 RSLCnano coupled via nano-ESI (NSI) to a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Proteins were denatured with 5 mM tris(2-carboxyethyl)phosphine (Sigma-Aldrich) at 60 °C and 900 rpm for 30 min. Disulfides were alkylated using 20 mM iodoacetamide (Sigma-Aldrich) at 22 °C and 750 rpm in darkness for 45 min. Protein subunits were purified using C18 pipet tips (Thermo Fisher Scientific), dried under vacuum (ScanVac, Lynge, Denmark), and digested with trypsin (Promega, enzyme/protein ratio = 1:25) overnight at 37 °C.
Resulting peptides were separated using a 75 μm I.D. × 150 mm PepMap Neo column packed with 2 μm C18 particles with 100 Å pores (Thermo Fisher Scientific) with 0.1% FA in H2O (A) and ACN (B) applying the following gradients: 1% B for 5 min, 1–30% B in 40 min, 30–60% B in 10 min, 99% B for 5 min, and 1% B for 20 min at 0.3 μL/min and 50 °C. The spray voltage for the NSI source probe was set at +1.4 kV, a probe heater temperature and capillary temperature at 350 and 250 °C, respectively, and an S-lens RF level at 60. Peptides were analyzed in a data-dependent MS/MS mode. Full scan was operated within a scan range of m/z 400–3000 at a resolution setting of 70,000, with an AGC target of 3e6, and a maximum injection time of 150 ms. Ten data-dependent scans were acquired at a normalized collision energy of 28 with a resolution setting of 17,500, an AGC target of 1e5, a maximum injection time of 150 ms, and a dynamic exclusion of 10 s. MS data were analyzed using Byonic (v. 3.11.3, Protein Metrics Inc., Cupertino). Peptides were quantified using Skyline (v. 20.2).
Results and Discussion
Retention and Selectivity Assessment
The retention characteristics of a mAb was evaluated on three HIC column chemistries, i.e., a conventional HIC phase based on butyl ligands (HIC-butyl), a bonded phase based on short strands of polymeric alkylamide, and a bonded phase based on long strands of a multialkylamide ligand. Table 1 provides an overview of stationary phase properties.
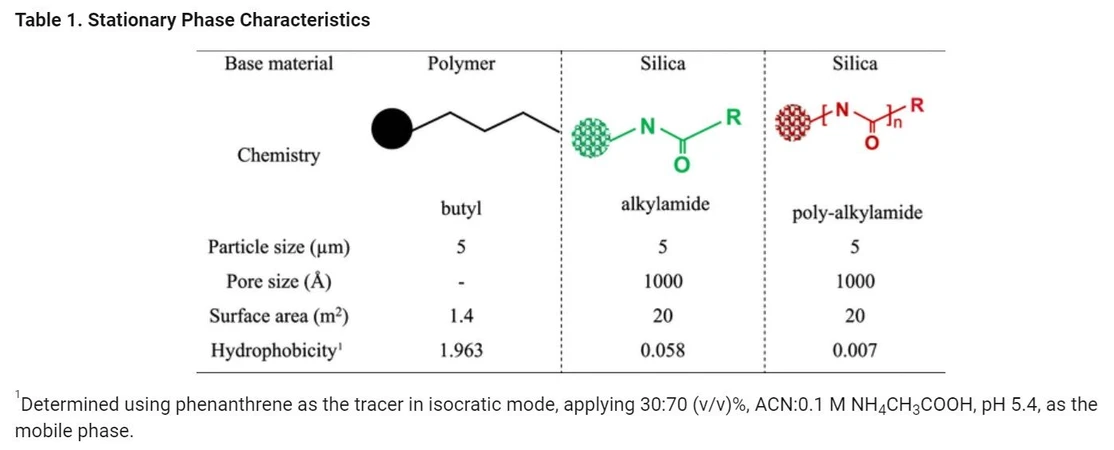 Anal. Chem. 2024, 96, 3, 1121-1128 - Table 1. Stationary Phase Characteristics.
Anal. Chem. 2024, 96, 3, 1121-1128 - Table 1. Stationary Phase Characteristics.
Figure 1 shows the peak profiles of the trastuzumab reference sample injected on the different columns by applying ammonium sulfate (Figure 1A) and sodium chloride (Figure 1B) salt systems...
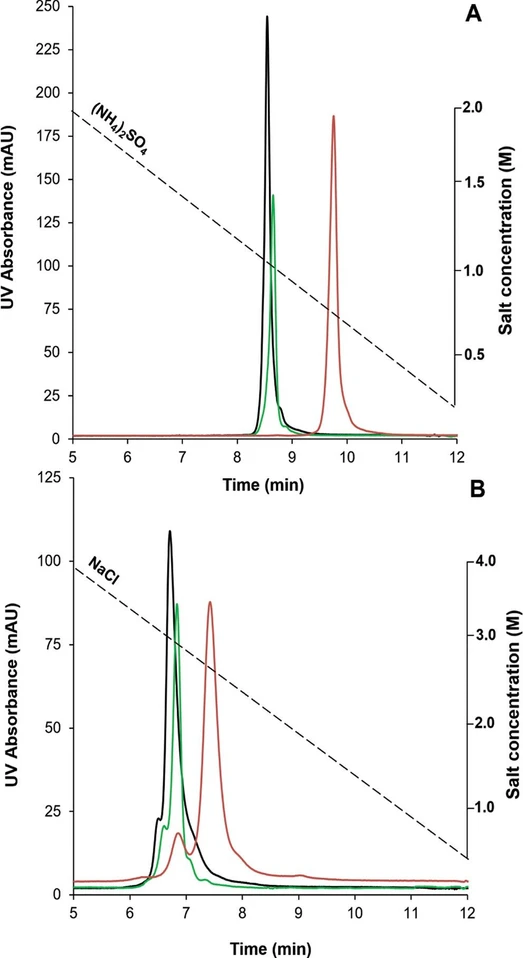 Anal. Chem. 2024, 96, 3, 1121-1128: Figure 1. Comparison of retention and selectivity obtained on different HIC columns for unstressed trastuzumab, applying inverse gradients of NH₄(SO₄)₂ from 2 to 0 M (A) and NaCl from 4 to 0 M (B) on a butyl phase (black profile), a bonded phase based on short strands of polymeric amide (green profile), and a covalently attached stationary phase based on a multiamide ligand (red profile). F = 1 mL/min, tG = 10 min, and the column oven was maintained at 30 °C.
Anal. Chem. 2024, 96, 3, 1121-1128: Figure 1. Comparison of retention and selectivity obtained on different HIC columns for unstressed trastuzumab, applying inverse gradients of NH₄(SO₄)₂ from 2 to 0 M (A) and NaCl from 4 to 0 M (B) on a butyl phase (black profile), a bonded phase based on short strands of polymeric amide (green profile), and a covalently attached stationary phase based on a multiamide ligand (red profile). F = 1 mL/min, tG = 10 min, and the column oven was maintained at 30 °C.
Optimizing the HIC Resolving Power
To generate an IgG sample with a substantial amount of oxidized amino acid residues, forced oxidation with hydrogen peroxide was pursued. To optimize the resolution between the resulting antibody variants, the effects of gradient span (Δc), gradient duration (tG), and column length were systematically assessed by injecting the reference standard before and after oxidation. As the determination of the peak width (even at half height) did not result in reliable performance metric, the valley-to-peak ratio (V/P) introduced by Christophe was used to evaluate performance, where V is the height of the valley between the critical peak pair and P is the height of the lowest peak, the lower the V/P, the better the separation. (26) Figure 3A shows the separation of the reference sample (black trace) and the separation of the antibody variant generated after forced oxidation is shown in Figure 3B (red trace) obtained on a 100 mm long column after optimizing Δc (3–0 M) and tG (30 min). When applying only a narrow gradient span from 2 to 0 M, the critical pair elutes close to the column hold-up time and the gradient duration is only utilized to a very small extent, resulting in an increase in V/P and decreasing the resolving power (data not shown). In the next step, the flow rate and gradient duration were systematically varied. Applying Δc of 3–0 M, a flow rate of 1 mL/min in combination with a tG of 30 min yielded a V/P of 0.40. Increasing tG led to a slight decrease in resolving power as the peak width increased linearly with gradient duration. On the other hand, reducing the flow rate to 0.5 mL/min led to an improved separation, yielding a V/P of 0.34. Operating at a lower flow rate results in a decrease in the mass-transfer contribution to the overall peak broadening, surpassing the impact of gradient steepness (tG/t0) on resolving power. To further optimize the separation, the effect of column coupling was assessed. Applying two coupled columns led to a further decrease in V/P, yielding 0.31, see Figure 3C (reference sample) and Figure 3D (trastuzumab after forced oxidation), respectively. Note that tG/t0 was scaled to column length, allowing an unbiased comparison with the performance obtained on one column. While one column displays limited resolution, novel oxidation variants become apparent in the coupled-column experiment.
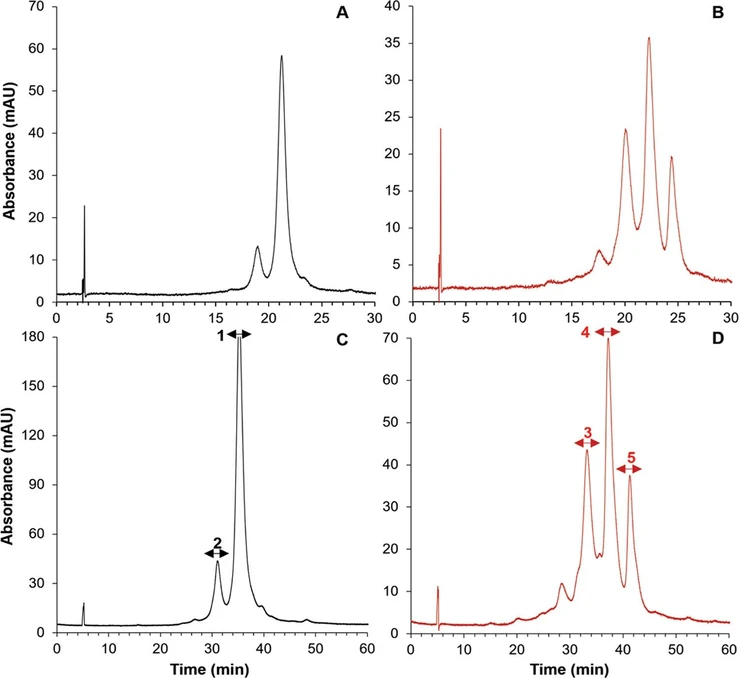 Anal. Chem. 2024, 96, 3, 1121-1128:Figure 3. Separation of the trastuzumab reference sample and trastuzumab variants after forced oxidation on one column (A, B) and two serially coupled columns (C, D). Separations were performed using the multialkylamide stationary phase, applying an inverse gradient of NaCl from 3 to 0 M, F = 0.5 mL/min, tG = 30 min (single column) and tG = 60 min (coupled columns). The fractions that were collected for subsequent RP-HPLC-MS/MS peptide mapping experiments, highlighted by the arrows (C, D).
Anal. Chem. 2024, 96, 3, 1121-1128:Figure 3. Separation of the trastuzumab reference sample and trastuzumab variants after forced oxidation on one column (A, B) and two serially coupled columns (C, D). Separations were performed using the multialkylamide stationary phase, applying an inverse gradient of NaCl from 3 to 0 M, F = 0.5 mL/min, tG = 30 min (single column) and tG = 60 min (coupled columns). The fractions that were collected for subsequent RP-HPLC-MS/MS peptide mapping experiments, highlighted by the arrows (C, D).
Conclusions
We highlight the critical role of phase systems for tuning chromatographic retention and selectivity. To effectively profile molecular mAb variants, a stationary phase chemistry offering complementary interaction modes, such as van der Waals interactions, H-bonding, π–π interactions, and steric interactions, presents abundant opportunities for unparalleled selectivity. In addition, protein–salt interactions can play a key role in achieving high-resolution HIC separation. While (NH4)2SO4 is widely regarded as the gold standard for HIC, chloride ions have been identified as a crucial factor in achieving selectivity, leading to the resolution between distinct molecular antibody variants. Although the linear solvent strength (LSS) plots for mAbs are relatively steep; retention in HIC is not based on an on–off mechanism. Consequently, the increasing column length has a profoundly positive effect on the resolution. This also indicates that the chromatographic performance can likely be further optimized when downscaling the particle diameter and increasing the operating pressure while ensuring protein conformation is not affected. Nonetheless, selectivity mediated by the ligand architecture and salt type should be prioritized during mAb variant analysis. Finally, our study also demonstrated that the site-specific deamidation of trastuzumab lowers the retention time. However, deamidated species can only be separated from the nondeamidated fraction in the presence of chloride ions. Also, site-specific oxidation induces protein unfolding, which, in turn, increases the retention time due to increased hydrophobic residue exposure.
- Selectivity and Resolving Power of Hydrophobic Interaction Chromatography Targeting the Separation of Monoclonal Antibody Variants. Raphael Ewonde Ewonde, Katharina Böttinger, Jelle De Vos, Nico Lingg, Alois Jungbauer, Christopher A. Pohl, Christian G. Huber, Gert Desmet, and Sebastiaan Eeltink. Analytical Chemistry 2024 96 (3), 1121-1128. DOI: 10.1021/acs.analchem.3c04011




