Evaluating the potential of hydrophilic interaction liquid chromatography for collagen peptide mapping analysis
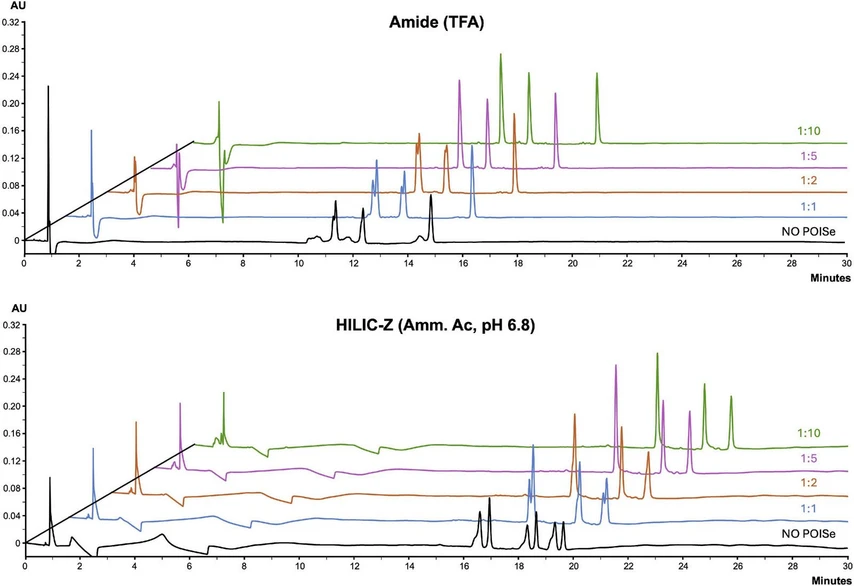
Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 6. Optimization of the ACN plug ratios for the injection, in POISe mode, of 4 µL from a 0.3 mg/mL solution of peptides #3, #8 and #15 to prevent peak splitting/deformation. Tests were performed both on the amide and zwitterionic columns with TFA and ammonium acetate as mobile phase additives, respectively. ACN-to-sample volume ratios were 1:1, 1:2, 1:5, and 1:10.
This study develops a systematic approach to optimizing hydrophilic interaction liquid chromatography (HILIC) for collagen peptide mapping analysis. Given collagen’s high proline hydroxylation, HILIC presents a promising alternative to reversed-phase chromatography. Sixteen model peptides were selected to evaluate chromatographic performance across various stationary and mobile phases, improving the understanding of peptide behavior in HILIC.
The study further examines sample injection techniques, including the Performance Optimizing Injection Sequence (POISe), to mitigate peak distortion caused by eluent strength mismatches. By introducing an acetonitrile plug before injection, peak shape and sensitivity were enhanced. Finally, coupling HILIC with mass spectrometry enabled peptide sequence coverage assessment and hydroxylation pattern identification, demonstrating HILIC’s potential for detailed collagen peptide analysis.
The original article
Evaluating the potential of hydrophilic interaction liquid chromatography for collagen peptide mapping analysis
Martina Lioi, Sara Tengattini, Valentina D'Atri, Gabriella Massolini, Simona Daly, Caterina Temporini, Davy Guillarme
Journal of Chromatography A, Volume 1738, 6 December 2024, 465473
https://doi.org/10.1016/j.chroma.2024.465473
licensed under CC-BY 4.0
Selected sections from the article follow. Formats and hyperlinks were adapted from the original.
1. Introduction
Collagen, the most abundant protein in the human body, offers an ideal opportunity to explore the potential of HILIC over traditional RPLC-based peptide mapping approaches. This interest arises from its high content of proline residues, which undergo enzymatic hydroxylation to form hydroxyproline [[11], [12], [13]], a PTM commonly associated with collagen. Hydroxylated proline residues, such as 4-hydroxyproline [14] and the less common 3-hydroxyproline [15,16], introduce polar functionalities that can interact favorably with hydrophilic stationary phases used in HILIC. These hydrophilic interactions can improve peptide retentivity, making HILIC a suitable platform for their analysis. Furthermore, the multiple hydroxylation sites in collagen peptides can complicate their chromatographic behavior. By exploiting the inherent hydrophilicity of collagen peptides and the distinctive retention mechanisms of HILIC, this analytical approach holds substantial potential to reveal the structural characteristics and modifications of collagen. Accurate analytical characterization is essential as even minor changes in hydroxylation patterns can affect protein folding and lead to severe biological consequences [[17], [18], [19], [20], [21]]. This is particularly relevant in the context of recombinant collagen production, where achieving the hydroxylation pattern is a critical quality attribute (CQA), and its evaluation could be of great importance. Traditionally, the degree of proline hydroxylation of collagen is assessed at the single amino acid level following acid hydrolysis and derivatization. Despite significant efforts to separate and quantify hydroxyproline at the amino acid level, the main drawback of this approach is the lack of sequence information [22].
This study presents a comprehensive and systematic approach aimed at developing a specific HILIC method for collagen peptide mapping, while also providing fundamental insights into peptide analysis. Sixteen model peptides, derived from in silico predicted tryptic peptides, were carefully selected to represent different physicochemical properties and structural motifs typical of collagen, with particular attention to proline hydroxylation. The research investigated various HILIC stationary phases under different mobile phase conditions. By examining the factors affecting peptide retention and selectivity in HILIC, the study aimed to identify the optimal separation conditions for collagen model peptides, and to enhance the understanding of the still underexplored chromatographic behavior of peptides in HILIC.
Particular attention was given to sample diluent and injection mode, known to be critical factors in HILIC. The solvent mismatch between sample diluent (predominantly aqueous) and mobile phase (mostly composed of acetonitrile) can significantly affect chromatographic performance, often leading to issues such as peak distortion, band broadening, and analyte breakthrough [23,24]. These problems arise because the higher elution strength of the sample can interfere with proper analyte retention on the hydrophilic stationary phase [25]. This is particularly critical for peptides and proteins analysis, where aqueous sample diluents are required to maintain solubility and prevent denaturation [26]. The Performance Optimizing Injection Sequence (POISe) provides an automated solution to these challenges. POISe involves the pre-injection of a defined volume of a weak solvent to stabilize the chromatographic system before the sample is introduced, thereby enhancing analytes interaction with the stationary phase during sample loading [[27], [28]]. This study systematically compares classical injection and POISe, focusing on its effects on the separation of collagen peptides. By optimizing the injection parameters, including the injection volume and solvent ratio, POISe can significantly improve the robustness and sensitivity of HILIC methods.
Finally, after optimizing the chromatographic separation, coupling the method with MS provided a thorough understanding of the method effectiveness in analyzing collagen-digested samples. This included a comprehensive evaluation of peptide sequence coverage and the ability to detect hydroxylation patterns, including the identification of positional isomers.
2. Materials and methods
2.2. Instrumentation, columns, chromatographic and MS conditions
Chromatographic analyses were conducted using a Waters ACQUITY UPLC H - Class System (Milford, USA) equipped with a quaternary solvent manager (QSM), an autosampler with a flow-through-needle (FTN), a thermostated column compartment, and a photodiode array UV detector (0.5 µL flow-cell). The injection system was configured to allow up to 50 µL injections by adding an external loop tubing. A 250 µL mixing chamber was included in the QSM to reduce baseline drifts. UV detection was performed at 214 nm with a sampling rate of 10 Hz and a filter time constant of 0.2 s. Median Baseline Filter (MBF) mode was systematically enabled during acquisition to limit baseline drift. Data acquisition, handling and instrument control were managed using Empower 3 software (Waters).
A variety of columns were tested in this study, including Waters Acquity UPLC BEH HILIC 130 Å, 1.7 µm (2.1 × 100 mm), Waters Acquity UPLC BEH Amide 130 Å, 1.7 µm (2.1 × 100 mm), Waters Atlantis BEH Z-HILIC 95 Å, 1.7 µm (2.1 mm x 100 mm), Agilent Poroshell 120 HILIC-Z 120 Å, 2.7 µm (2.1 × 100 mm), Daicel DCpak PMPC 3 µm (4.6 mm x 150 mm), Agilent Poroshell 120 HILIC - OH5 2.7 µm (2.1 × 100 mm), Waters Acquity UPC2 Torus 2-PIC, 130 Å, 1.7 µm (3 mm x 100 mm), Waters Acquity UPC2 BEH 2-EP, 1.7 µm (3 mm x 100 mm), Daicel DCpak PTZ, 3 µm (2.1 × 150 mm), Waters Acquity UPC2 Torus Diol, 130 Å, 1.7 µm (3 mm x 100 mm) and Fortis HILIC Diol 1.7 µm (2.1 mm x 50 mm).
A multiple injection mode was employed in this study. The ACQUITY FTN autosampler was configured to perform a multi-step (sequenced) injection program using the Auto-Additions feature in Empower. This setup allows for automated programming of injections, where a specified volume of the sample is aspirated, followed by a specified volume of solvent. ACN was selected as solvent for these injections.
UHPLC - HRMS analyses were carried out on an Ultimate 3000 LC System (Thermo Scientific™ San Jose, CA, USA) equipped with quaternary pump, thermostated column compartment, and autosampler. This system was coupled to a Q Exactive™ Orbitrap with a heated electrospray ionization (HESI) source. Acquisition was performed in positive ion mode. Spray voltage was set at 3.7 kV, sheath gas flow at 30 and aux gas flow at 15. The capillary temperature was 320 °C, the mass range was 350–1800 m/z, AGC was 3 × 106, resolution was 7 × 104, and maximum injection time 120 ms. Data were acquired in data-dependent acquisition mode, and the fragmentation mass spectra were obtained by high-energy collisional dissociation (HCD) with dynamic exclusion (repeat count of 3, exclusion duration of 10 s), AGC of 1 × 105, maximum injection time of 200 ms, and isolation window of 2.0 m/z. Normalized collision energy was set to 28 %.
MaxQuant (v2.6.2.0) was used for peptide mapping analysis. Mass spectra were searched against the human collagen alpha-1(II) chain sequence obtained from UniProt (CO2A1_HUMAN, P02458). Search parameters included trypsin as the enzyme, with a maximum of two missed cleavages allowed. The minimum peptide length was set to 5 amino acids and the maximum peptide mass to 4600 Da Carbamidomethylation (+57.0215 Da) was considered as a fixed modification and hydroxyproline (+15.9949 Da) as a variable modification.
3. Results and discussion
3.2. Analysis of model peptides: chromatographic method development and fundamental studies on peptides in HILIC
A sample containing the sixteen model peptides was prepared at a final concentration of 1.6 mg/mL (0.1 mg/mL each), by diluting with water to simulate the aqueous environment of tryptic digestion. Preliminary screening involved testing various classical and more exotic stationary phases, as listed in Section 2.2. Among the most exotic columns that were tested in this work, we can cite some columns commonly used in supercritical fluid chromatography (SFC), such as Torus 2-PIC (2-picolylamine ligand), BEH 2-EP (2-ethypyridine ligand) or Torus Diol, which possess a polar character. In addition, two columns from Daicel, namely DCpak PMPC (Poly (2-methacryloyloxyethyl phosphorylcholine) bonded to silica gel) and DCpak PTZ (Poly N-(1H-tetrazole-5-yl)-methacrylamide bonded to a silica gel carrier) were also tested. Unfortunately, most of the HILIC columns tested in this study offered some disappointing performance, with either very poor selectivity, no elution of peptides or complete lack of retention (data not shown). Therefore, only three columns - Acquity BEH Amide 130 Å, 1.7 µm (2.1 × 100 mm), Acquity BEH HILIC 130 Å, 1.7 µm (2.1 × 100 mm) and Poroshell 120 HILIC-Z 120 Å, 2.7 µm (2.1 × 100 mm) - were ultimately selected for in-depth studies.
Each stationary phase was evaluated under two distinct mobile phase conditions. In the first condition, TFA was used as an ion-pairing agent, added to both water (mobile phase A) and ACN (mobile phase B). Although TFA is generally not recommended for MS analysis, due to its potential to suppress ionization and affect ion stability in ionization sources, a low concentration (0.05 % in mobile phase A and 0.04 % in B) was used in this study to promote ion-pairing and enhance efficiency, with minimal sensitivity reduction. The second condition employed a mobile phase containing 20 mM ammonium acetate at pH 6.8 (mobile phase A) and ACN (mobile phase B). During this part of the study where the goal was to assess peptide elution orders and overall resolving power across different stationary and mobile phase conditions, the injection volume was set at 0.5 µL to limit the impact of sample diluent on peak shape. A wide generic gradient - where mobile phase B varied from 95 to 40 % over 30 min - was used, keeping in mind the final application to complex digested samples of collagen, which include numerous peptides that may elute outside the range of the sixteen model peptides.
Fig. 1 provides a comprehensive overview of the separation achieved for all sixteen model peptides, while Fig. 2 focuses specifically on peptide #2 and its hydroxylated variants. Both figures include data from all three columns tested with the two distinct mobile phases (TFA and ammonium acetate at pH 6.8). To determine the elution orders, peptides were individually injected from a 0.1 mg/mL solution prepared by diluting the stock solution with water. Notably, the amide column combined with TFA emerged as the most effective condition, successfully resolving fifteen out of the sixteen peptides, as shown in Fig. 1. Fig. 2 further illustrates that this setup was particularly effective in differentiating peptide #9 from peptide #10, resolving the 3-hydroxyproline and 4-hydroxyproline on the same proline residue. The only instance of coelution occurred between peptide #7 and peptide #10 due to 4-hydroxyproline on an adjacent proline residue. TFA-based mobile phases generally provided superior selectivity and better peak shapes compared to ammonium acetate. Regarding the separation of hydroxylated variants, the amide column performed best, followed by the zwitterionic and the bare silica. All the three stationary phases were able to distinguish peptide #9 from peptide #10, but coelutions became progressively more significant, with bare silica resolving only two out of the five modified peptides related to peptide #2 (Fig. 2). On the other hand, the use of amide and bare silica columns with ammonium acetate proved unreliable, due to insufficient sensitivity with a 0.5 µL injection volume. To obtain suitable chromatograms for these columns, as displayed in Figs. 1 and 2, the injection volume had to be increased to 3 µL. Furthermore, these setups exhibited several issues such as multiple peaks eluted before the main ones and inconsistent elution orders, particularly with the bare silica column, complicating data interpretation. The only column that produced reliable results with ammonium acetate was the zwitterionic, which resolved eleven out of sixteen peptides and demonstrated good sensitivity with the 0.5 µL injection, consistent with the performance observed with TFA-based conditions.
 Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 1. Chromatographic separation of sixteen collagen model peptides using amide, bare silica and zwitterionic stationary phases. The mobile phases were water + 0.05 % TFA in A and ACN + 0.04 % TFA in B (blue) and 20 mM ammonium acetate, pH 6.8 in A and ACN in B (orange).
Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 1. Chromatographic separation of sixteen collagen model peptides using amide, bare silica and zwitterionic stationary phases. The mobile phases were water + 0.05 % TFA in A and ACN + 0.04 % TFA in B (blue) and 20 mM ammonium acetate, pH 6.8 in A and ACN in B (orange).
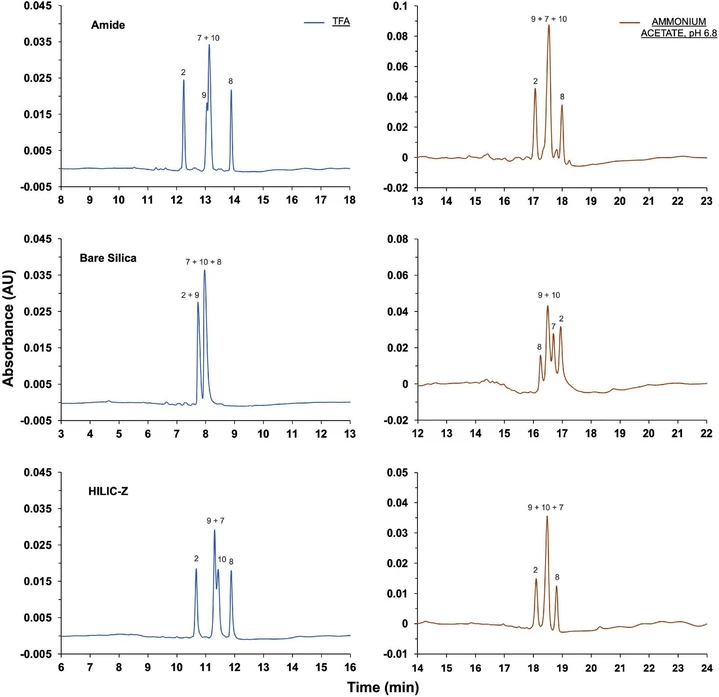 Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 2. Chromatographic separation of peptide #2 (EGSPGADGPPGR) and its hydroxylated versions: peptide #7 (EGSPGADGP4-HypGR), peptide #8 (EGSPGADG3-Hyp4-HypGR), peptide #9 (EGSPGADG3-HypPGR) and peptide #10 (EGSPGADG4-HypPGR). Analyses were carried out on the amide, bare silica and zwitterionic stationary phases. The mobile phases were water + 0.05 % TFA in A and ACN + 0.04 % TFA in B (blue) and 20 mM ammonium acetate, pH 6.8 in A and ACN in B (orange).
Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 2. Chromatographic separation of peptide #2 (EGSPGADGPPGR) and its hydroxylated versions: peptide #7 (EGSPGADGP4-HypGR), peptide #8 (EGSPGADG3-Hyp4-HypGR), peptide #9 (EGSPGADG3-HypPGR) and peptide #10 (EGSPGADG4-HypPGR). Analyses were carried out on the amide, bare silica and zwitterionic stationary phases. The mobile phases were water + 0.05 % TFA in A and ACN + 0.04 % TFA in B (blue) and 20 mM ammonium acetate, pH 6.8 in A and ACN in B (orange).
3.4. Application to collagen tryptic digestion: HILIC-MS/MS peptide mapping
The two most promising columns, Acquity UPLC BEH Amide 130 Å, 1.7 µm (2.1 × 100 mm) and Poroshell 120 HILIC-Z 120 Å, 2.7 µm (2.1 × 100 mm), along with their respective chromatographic conditions previously detailed (TFA on the amide column and ammonium acetate on the zwitterionic column), were selected for collagen peptide mapping analysis. A 1 mg/mL solution of trypsin-digested human collagen (type II) was employed for this purpose, and analyses were carried out on a Q-Exactive Orbitrap LC-MS/MS system.
Injection of 8 µL in POISe mode on the amide column enabled 63.8 % amino acid sequence coverage to be achieved, with 78 unique peptide sequences identified. In comparison, the zwitterionic column yielded the identification of 66 unique sequences from the same sample, covering 60.7 % of the collagen sequence. The list of identified peptides is provided in the Supplementary Information. Notably, among the peptides identified, 70 from the amide and 57 from the zwitterionic were hydroxylated, indicating the prevalence of this modification in the collagen structure. Overall, considering both unmodified and differently hydroxylated peptides, 138 species were identified on the amide column and 104 on the zwitterionic. Interestingly, all six unmodified peptides selected as standards in the first part of this work were also identified in the sample, thereby validating their representativeness. The observed hydroxylation pattern slightly varied from our theoretical speculations, except for peptide #11 (GEVGPPGPAGSAGAR) and peptide #16 (GLPGTPGTDGPK), which matched the expected number and positions of hydroxyprolines.
Sequence coverage increased by approximately 15 % when the injection volume increased from 0.5 to 8 µL, confirming the need to maximize sample injection for optimal results. The use of classical injection or POISe mode did not consistently affect amino acid sequence coverage. However, the extracted ionic current of an illustrative peptide, namely GEVGPPGPAGSAGAR (m/z = 648.32057, z = 2), reveals that peak distortion occurred in the complex tryptic sample, not just in the standard mixture (Fig. 7A). POISe injection effectively mitigated the impact of solvent mismatch, improving peak shape and resolution (Fig. 7B), as also previously demonstrated. Furthermore, the use of POISe enhances signal intensity, resulting in better-defined peaks. This improvement in intensity is of particular importance for the detection of low-abundant peptides, which may otherwise be missed in a complex sample matrix. These advantages are demonstrated in Fig. 7, showing the superior peak quality and intensity achieved through POISe injection.
 Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 7. Extracted ion current for peptide GEVGPPGPAGSAGAR (m/z = 648.32057, z = 2) in a 1 mg/mL human collagen type II tryptic digest, analyzed using both amide and zwitterionic stationary phases. The results obtained from 8 µL injected in classical mode (upper panel) and POISe mode (lower panel) are visually compared.
Journal of Chromatography A, Volume 1738, 6 December 2024, 465473: Fig. 7. Extracted ion current for peptide GEVGPPGPAGSAGAR (m/z = 648.32057, z = 2) in a 1 mg/mL human collagen type II tryptic digest, analyzed using both amide and zwitterionic stationary phases. The results obtained from 8 µL injected in classical mode (upper panel) and POISe mode (lower panel) are visually compared.
4. Conclusion
This study successfully demonstrates the utility of HILIC for collagen peptide mapping analysis, providing a viable alternative to traditional RPLC-based methods. By thoroughly evaluating chromatographic conditions, including stationary phases, mobile phases, and sample injection modes, we have identified effective strategies for the analysis of collagen peptides, characterized by the extensive modification of proline residues to polar hydroxyproline. These findings also provide a broader understanding of peptide performance on HILIC.
Two stationary phases, amide and zwitterionic, were found to be particularly well suited to this task. The amide column, with the addition of 0.05 % TFA in water and 0.04 % in ACN, demonstrated superior resolution, especially in the separation of hydroxylated peptides and positional isomers. On the other hand, the zwitterionic column proved to be optimal for peak shape, even when injecting large sample volumes. The study also highlights the impact of sample diluent and injection mode, emphasizing the importance of managing solvent mismatch and optimizing injection strategies to enhance chromatographic performance. The addition of a weak solvent plug (acetonitrile) prior to sample injection significantly improved peak shape and prevented peak splitting and distortion, which were initially observed due to the fully aqueous nature of sample diluent in tryptic digestions. These phenomena, related to the incompatibility of solvent strengths between sample diluent and mobile phases, were observed for both standard collagen peptides and tryptic peptides. POISe injection allowed the successful maximization of injection volume (up to 8 μL), increasing sensitivity, while maintaining optimal peak shape. These observations on peptides are important, as the effects of POISe injection have so far been evaluated only for small molecules and intact proteins. Coupled with MS, both HILIC columns facilitated detailed analysis of hydroxylation patterns, crucial for understanding the structural and functional properties of collagen. Both columns effectively resolved peptides with different numbers of hydroxyprolines and located at different positions (positional isomers). The resolving power of HILIC, combined with the ability of MS to localize the modified proline residue, underlines its potential as a valid alternative to the traditionally employed RPLC.




