Oligos Made Easy - Part 3: IP-RP

- Photo: KNAUER: Oligos Made Easy - Part 3: IP-RP
- Video: KnauerHPLC: The OLIGOSCALER by KNAUER presented by Marvin Schäk
Mastering Ion-Pair Reversed-Phase Chromatography for Oligonucleotide Analysis
Why Purity Is Critical in the Oligonucleotide Era
Oligonucleotides — short, synthetic strands of DNA or RNA — are central to modern biotechnology. They power gene therapies, advanced diagnostics, and cutting-edge molecular research. However, before these molecules can be used in clinical or research settings, their purity and structural integrity must be thoroughly verified. High-resolution chromatographic analysis plays a crucial role in ensuring quality and safety.
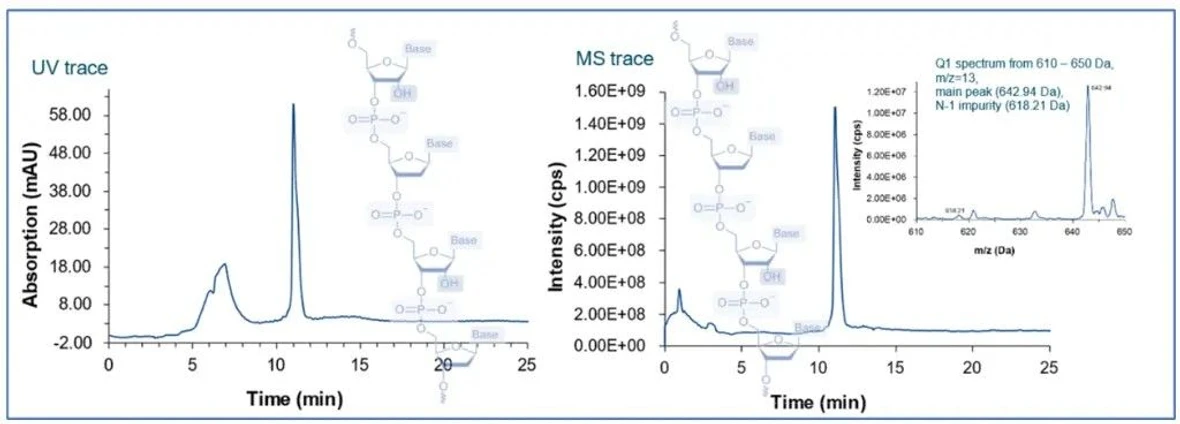 KNAUER: Figure 1 - Separation Example of Oligonucleotides.
KNAUER: Figure 1 - Separation Example of Oligonucleotides.
Understanding Ion-Pair Reversed-Phase (IP-RP) Chromatography
For scientists involved in oligonucleotide synthesis and characterization, Ion-Pair Reversed-Phase Chromatography (IP-RP) is one of the most effective analytical tools available.
Unlike separation techniques that rely solely on charge differences, IP-RP combines:
- Ionic interactions
- Hydrophobic interactions
This dual retention mechanism enables exceptional resolution — even for oligonucleotides that differ by only a single nucleotide or subtle chemical modification. Whether assessing synthesis efficiency or purifying therapeutic-grade material, IP-RP provides reliable, reproducible separations.
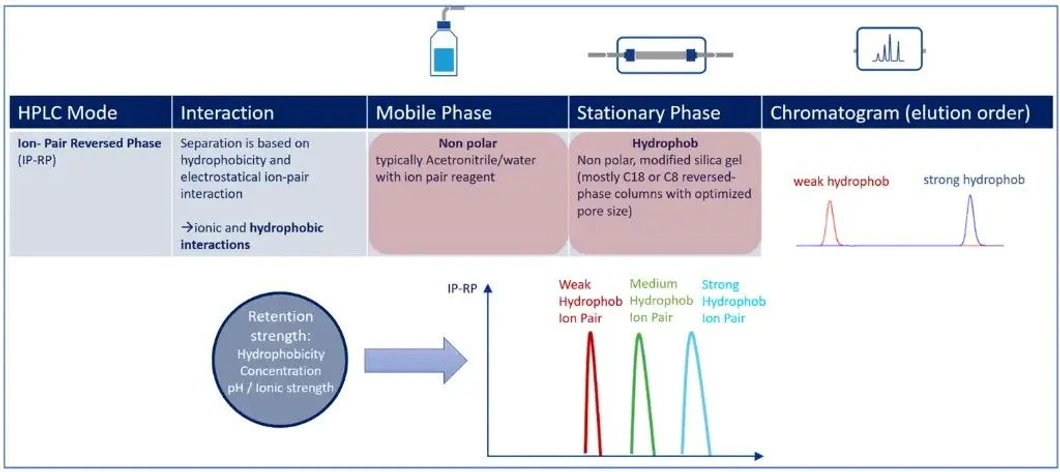 KNAUER: Figure 2 - Overview of IP-RP Mode prinicple.
KNAUER: Figure 2 - Overview of IP-RP Mode prinicple.
The Principle Behind Ion-Pairing
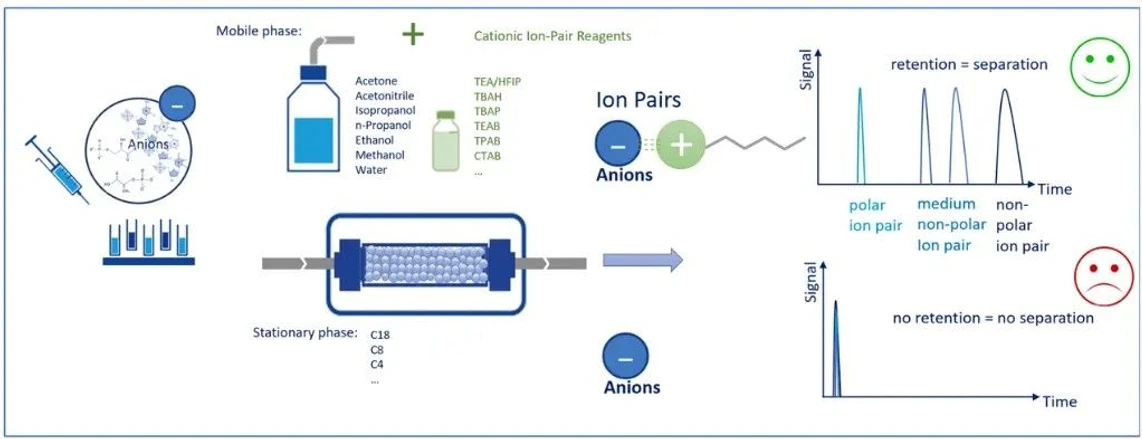
Oligonucleotides possess a highly negatively charged phosphate backbone. In a traditional reversed-phase system, such hydrophilic, charged molecules would show little retention.
IP-RP overcomes this limitation by introducing a positively charged ion-pairing reagent into the mobile phase. The cation associates with the negatively charged oligonucleotide, forming a transient ion pair with increased hydrophobic character. This modified complex can now interact with the hydrophobic stationary phase.
Retention strength depends on:
- The hydrophobicity of the ion-pair reagent
- The strength of ionic interaction
- The applied organic gradient
During gradient elution (commonly using acetonitrile), analytes are released in a controlled manner according to size, sequence, and chemical modifications.
 KNAUER: Figure 3 - Ion-Pair Reversed Phase Principle.
KNAUER: Figure 3 - Ion-Pair Reversed Phase Principle.
Choosing the Right Ion-Pair Reagent
Successful IP-RP separation depends heavily on selecting an appropriate ion-pair reagent. Two key factors determine retention behavior:
- Alkyl chain length
- Charge density
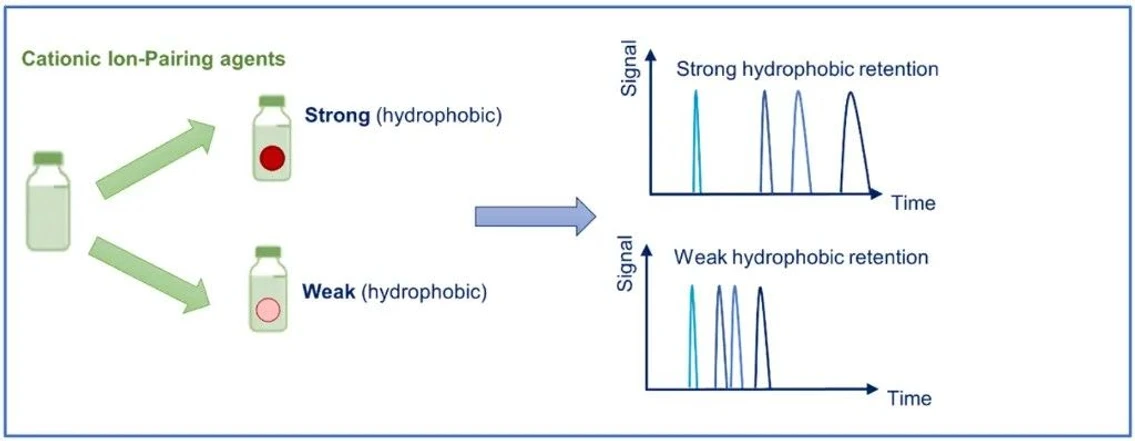 KNAUER: Figure 4 - Ion-Pair Classification.
KNAUER: Figure 4 - Ion-Pair Classification.
Strong Cationic Ion-Pair Reagents
Ion-pair reagents with longer alkyl chains provide stronger hydrophobic interactions and increased retention. A widely used example is tetrabutylammonium (TBA⁺), frequently applied for strongly retained anionic analytes.
These reagents are particularly useful when high retention strength is required.
 KNAUER: Table 1 - Strong Cationic Ion-Pair Reagents.
KNAUER: Table 1 - Strong Cationic Ion-Pair Reagents.
Volatile (Weaker) Ion-Pair Reagents
Short-chain ion-pair reagents generate weaker retention but offer a significant advantage: compatibility with LC–MS detection.
Triethylammonium (TEA⁺) is among the most commonly used reagents for oligonucleotide analysis, particularly when mass spectrometric detection is involved. Other options, such as diisopropylethylammonium (DIEA⁺), are also widely applied, often combined with HFIP to enhance MS performance.
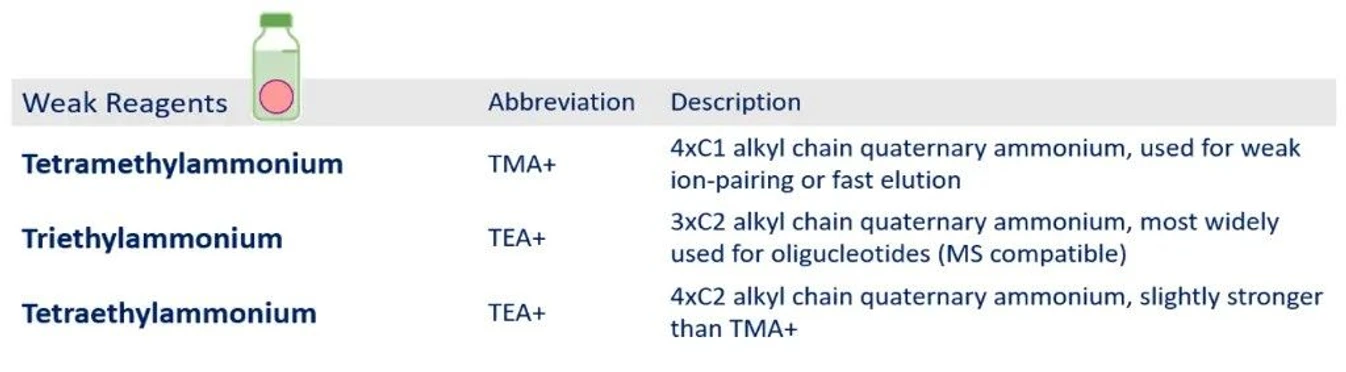 KNAUER: Table 2 - Weak Cationic Ion-Pair Reagents.
KNAUER: Table 2 - Weak Cationic Ion-Pair Reagents.
Building an IP-RP Method for Oligonucleotides
Developing a robust IP-RP method follows a logical sequence:
 KNAUER: Table 3 - IP-RP Method Step Procedure for Oligos.
KNAUER: Table 3 - IP-RP Method Step Procedure for Oligos.
1. Characterize the Analyte
Oligonucleotides typically contain 10–50+ nucleotides and exhibit high negative charge density. This necessitates a suitable cationic partner such as TEA⁺ or DIEA⁺.
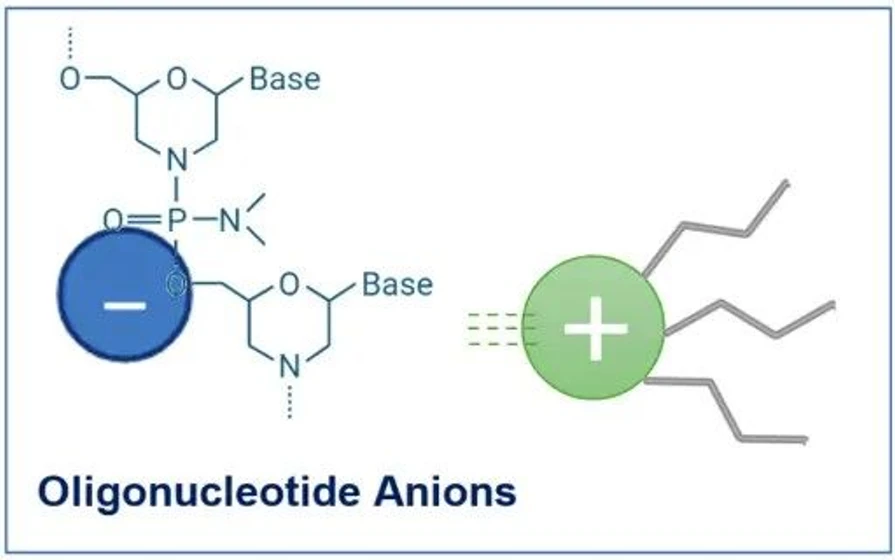 KNAUER: Figure 5 - Cationic Ion-Pair Principle for Oligonucleotides.
KNAUER: Figure 5 - Cationic Ion-Pair Principle for Oligonucleotides.
2. Select the Column
Use a reversed-phase column with:
- Large pore size (≈300 Å)
- Appropriate internal diameter
These parameters support the so-called “ON–OFF” retention mechanism — a bind-and-elute behavior where analytes are strongly retained until the gradient triggers rapid elution.
3. Optimize the Mobile Phase
A typical starting condition includes:
- 10 mM TEA buffer at pH ~7
- Acetonitrile as organic modifier
Gradient programs from approximately 5% to 50% organic solvent over 15–30 minutes are commonly effective for complex oligonucleotide mixtures.
4. Adjust Retention Strength if Needed
To increase retention:
- Raise ion-pair concentration (up to ~100 mM if required)
- Select a more hydrophobic ion-pair reagent
5. Validate the Method
Ensure robustness, sensitivity, and reproducibility by carefully evaluating:
- UV chromatograms
- MS traces
- Peak shape and resolution
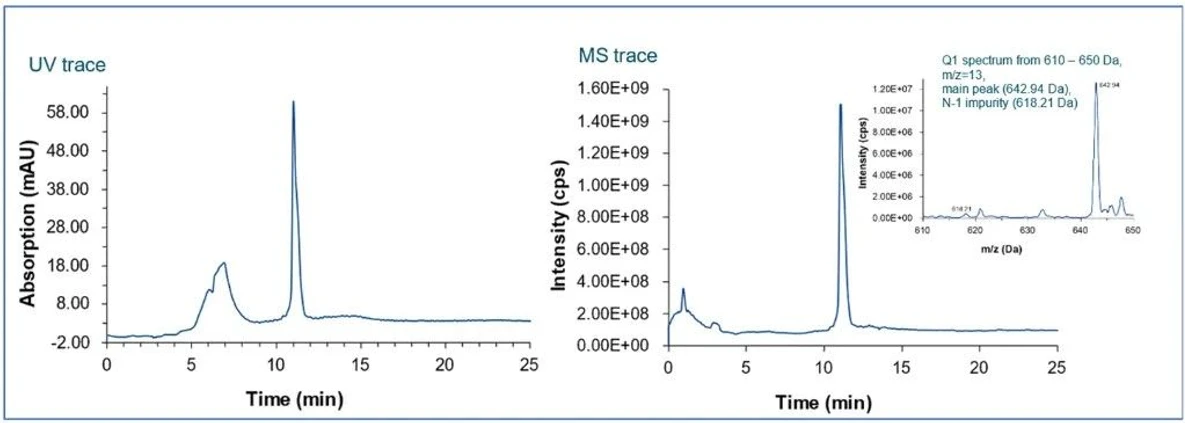 KNAUER: Figure 6 - UV-MS traces of IP-RP Oligonucleotide Analysis.
KNAUER: Figure 6 - UV-MS traces of IP-RP Oligonucleotide Analysis.
Why IP-RP Is Considered the Gold Standard
Ion-Pair Reversed-Phase Chromatography uniquely combines the resolving power of reversed-phase LC with the ability to retain highly polar, charged biomolecules.
Its advantages include:
- Tunable selectivity
- Compatibility with UV and MS detection
- Scalability from analytical to preparative workflows
- Applicability from research to pharmaceutical production
By transforming highly charged oligonucleotides into hydrophobic ion-pair complexes, IP-RP enables precise and reproducible separations that would otherwise be impossible in conventional reversed-phase systems.
For these reasons, IP-RP is widely regarded as the benchmark technique for oligonucleotide analysis.
Final Thoughts
Mastering IP-RP is less intimidating than it may seem. A successful method comes down to:
- Selecting the right ion-pair reagent
- Choosing a suitable column
- Optimizing gradient conditions
- Fine-tuning temperature and concentration parameters
With thoughtful method development, IP-RP delivers sharp peaks, clean separations, and reliable performance — fully compatible with modern mass spectrometry workflows and ready to meet the demands of advanced oligonucleotide analysis.